CutePeaks
CutePeaks is a user-friendly cross platform Sanger Trace file viewer written in C++ with the framework Qt5. It can handle ABIF and FSA file and provide some improvement like sequence editing, regular expression pattern finder and export as vector image. Cutepeaks has been packages for Windows, Mac and Linux.
Installation
Windows
cutepeaks auto installer
cutepeaks standalone zip
MacOSX
Linux
Linux binary is available as AppImage. Download the AppImage from here. Run it as follow:
chmod +x cutepeaks-0.2.0-linux-x86_64.appimage
./cutepeaks-0.2.0-linux-x86_64.appimage
From source code
Install Qt ≥ 5.7
From website: Download Qt ≥ 5.7 from https://www.qt.io/. Don’t forget to check QtChart module during installation.
From Ubuntu: Qt 5.7 is not yet available with Ubuntu. But you can add a PPA to your software system.
sudo apt-get install build-essential
sudo apt-get install qt5-default libqt5charts5-dev
From Fedora: Qt 5.7 is available.
sudo dnf install qt5-qtbase-devel qt5-qtcharts-devel
Compile CutePeaks
Be sure you have the correct version of Qt (≥ 5.7) by using qmake –version.
qmake cutepeaks.pro
make
sudo make install
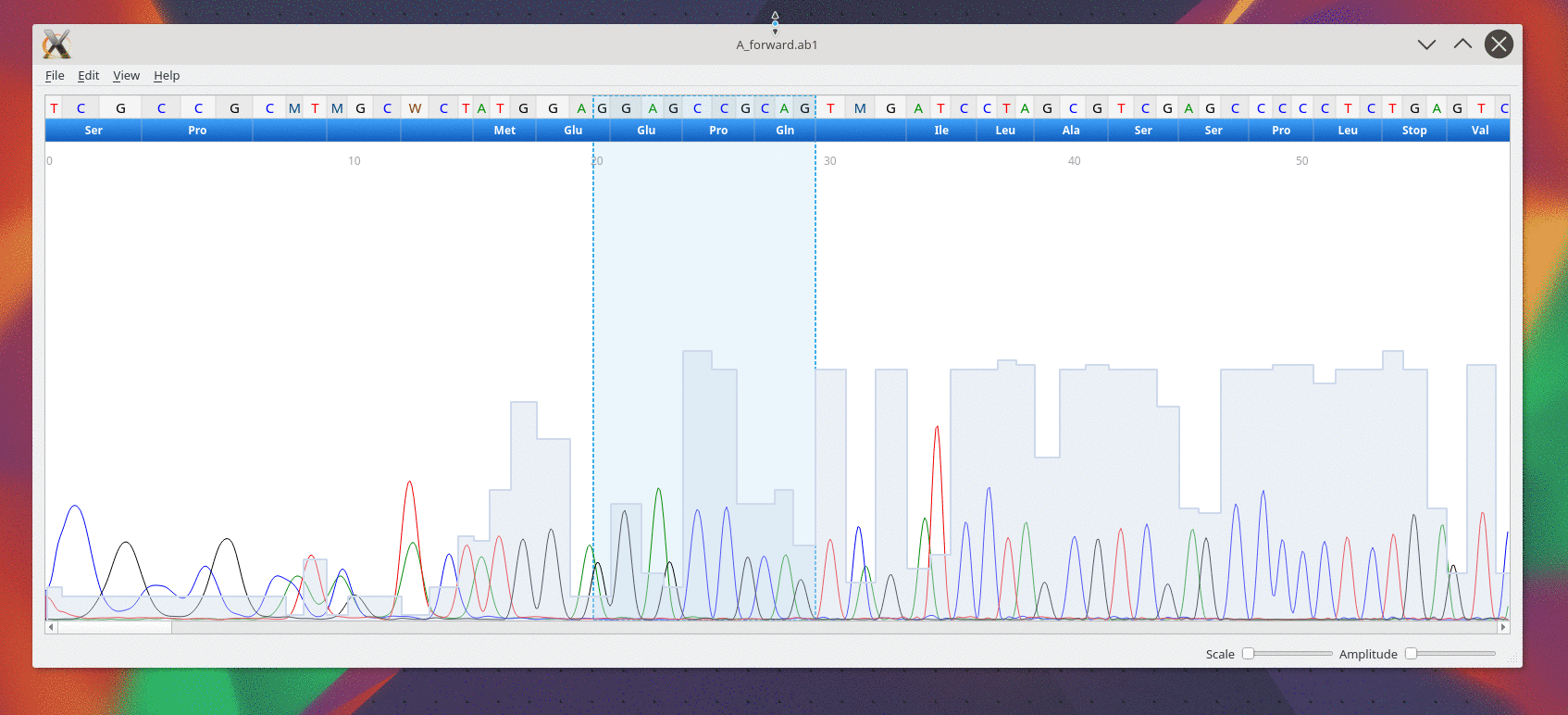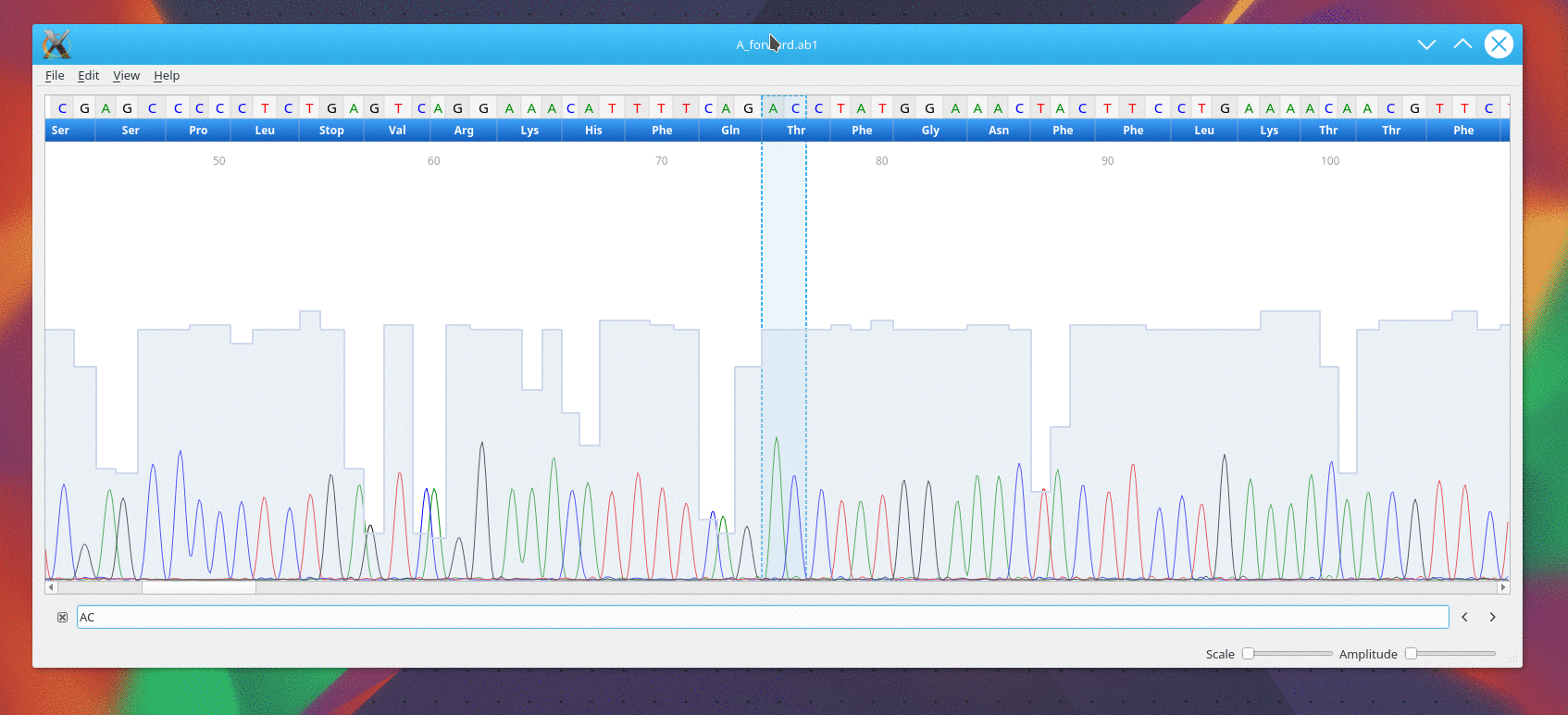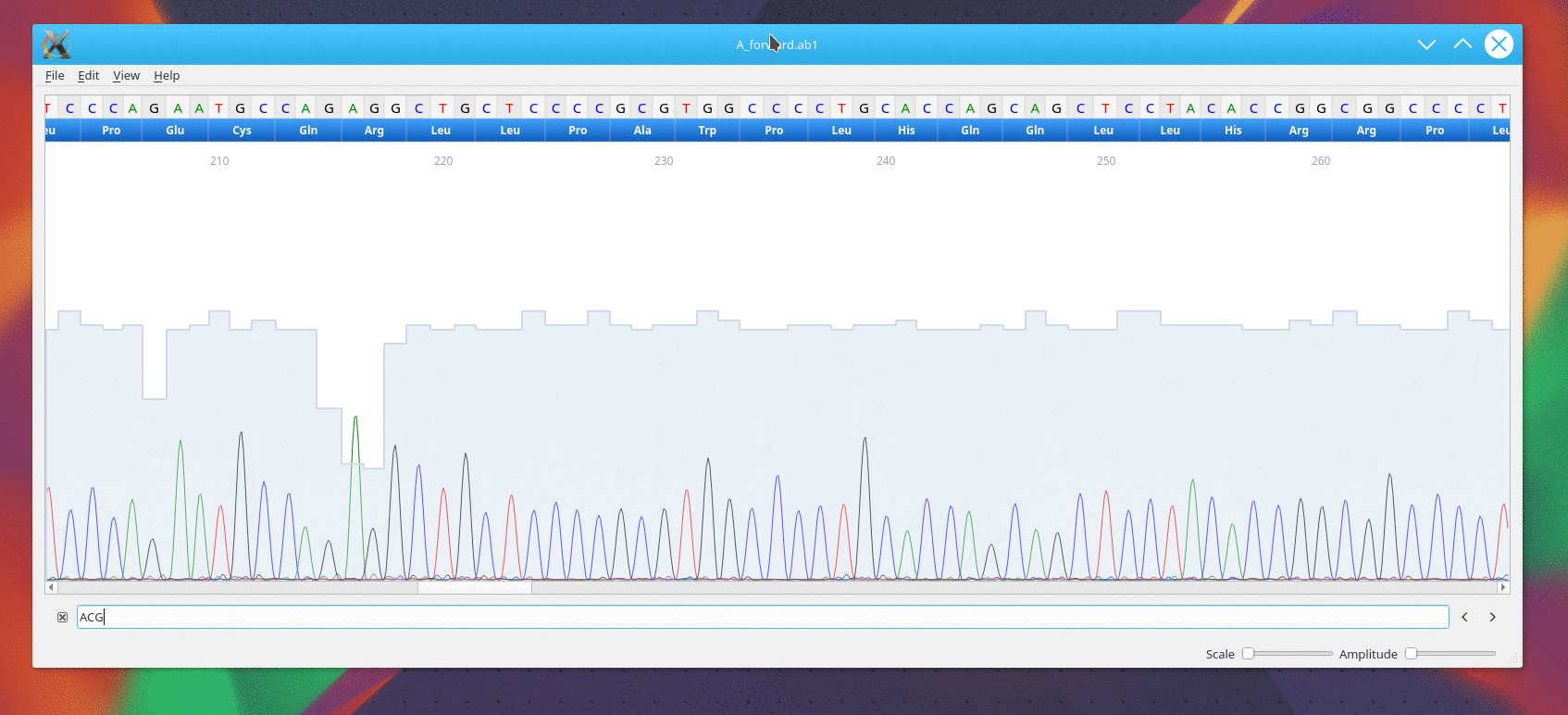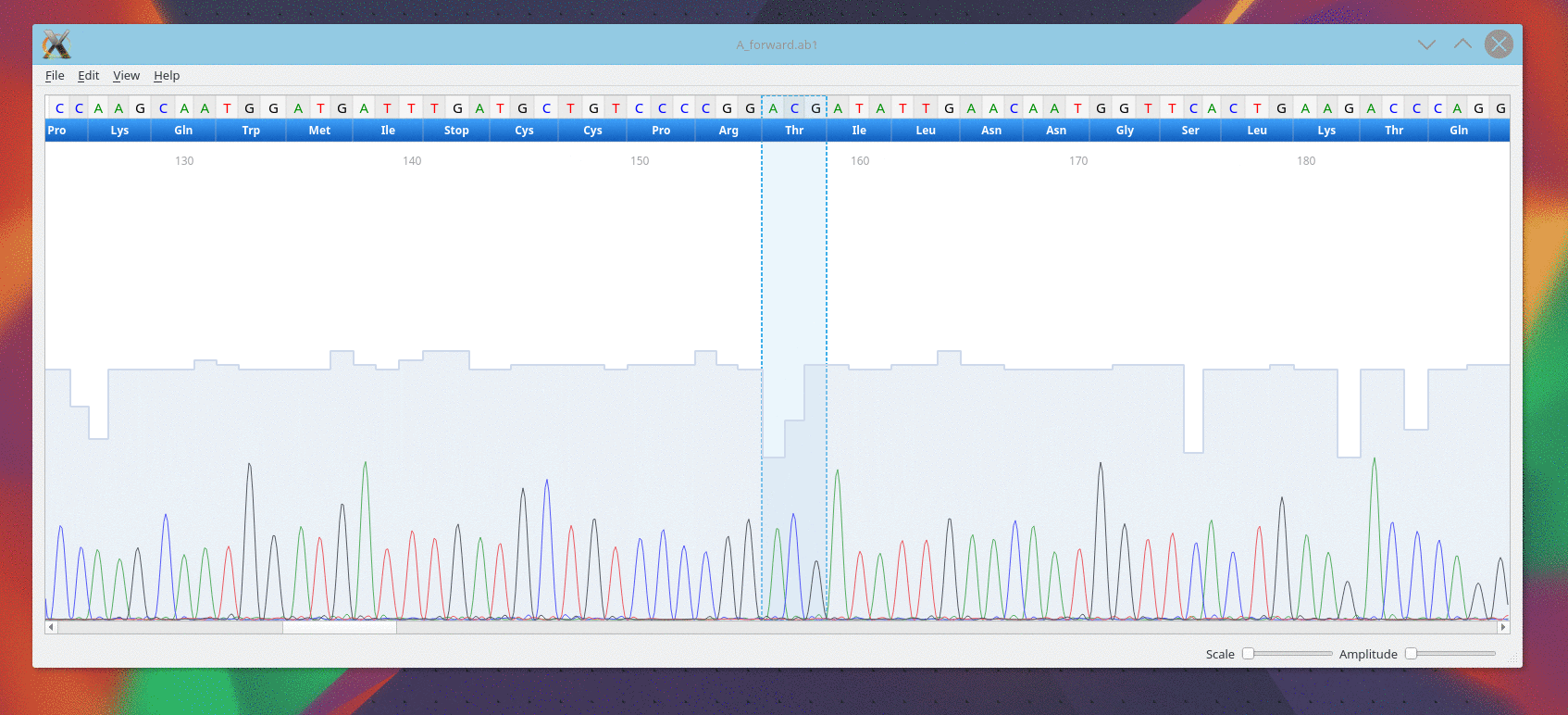
Usage
CutePeaks supports following trace file formats:
- *.ab1 : Applied Biosystems Genetic Analysis
- *.scf : Standard Chromatogram Format
Example file
An example file is available here for testing purpose A_forward.ab1